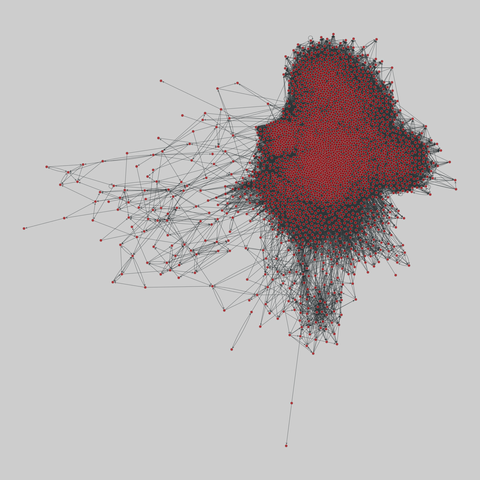
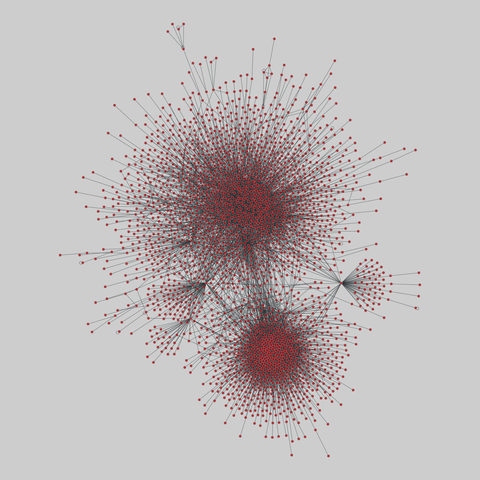
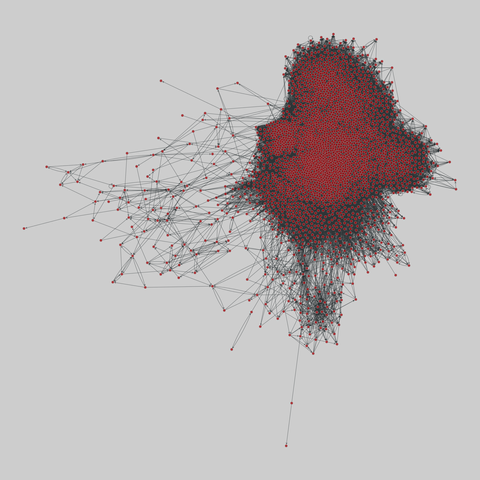
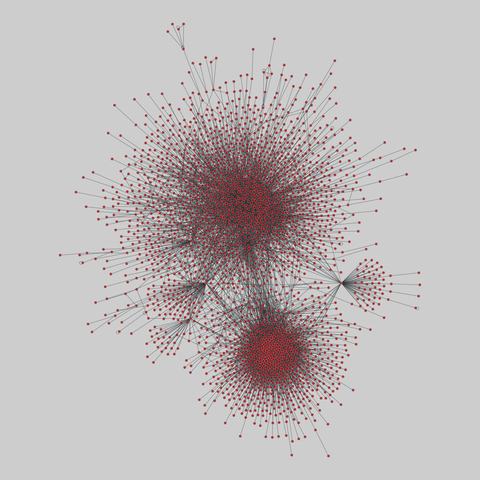
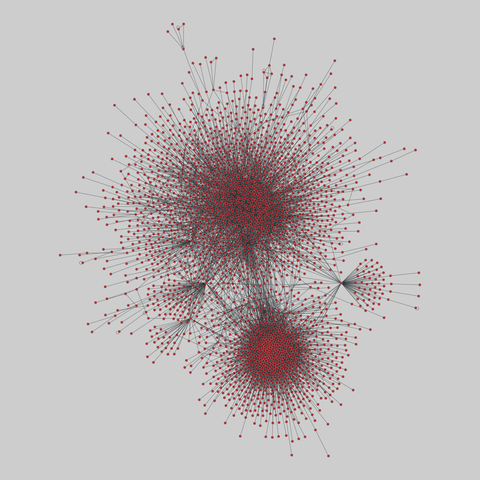
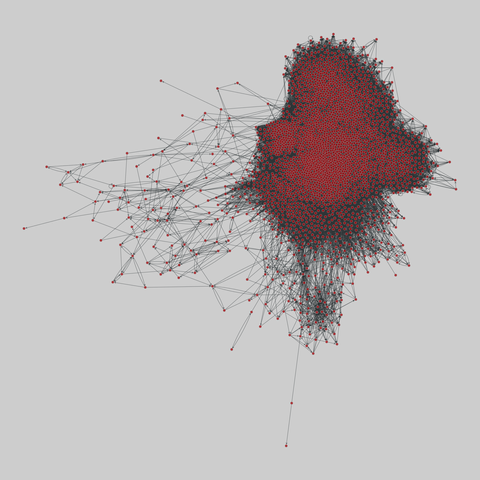
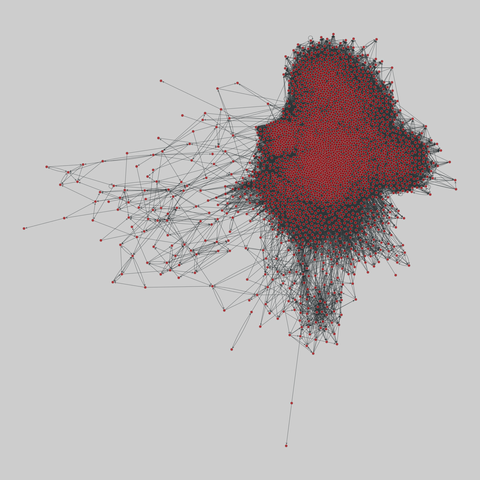
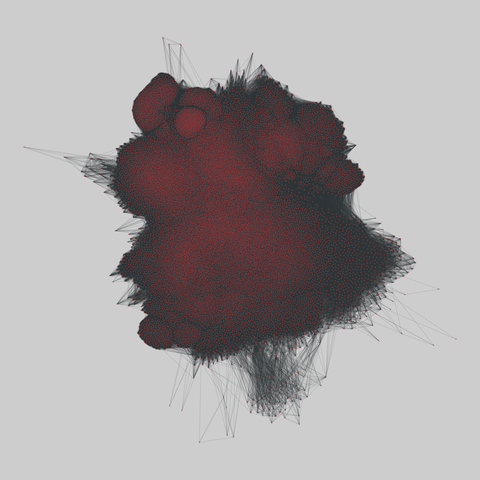
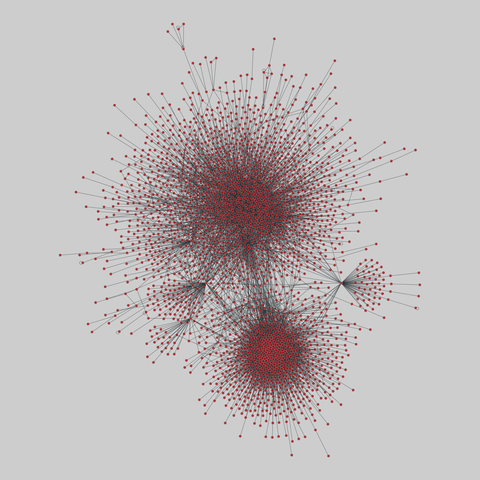
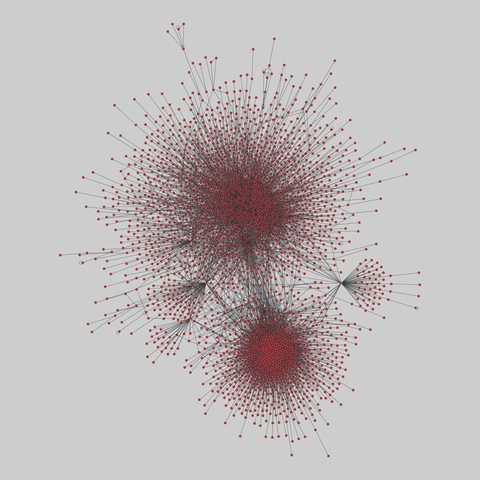
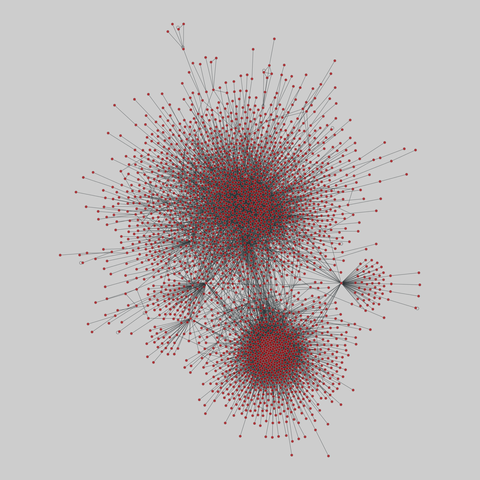
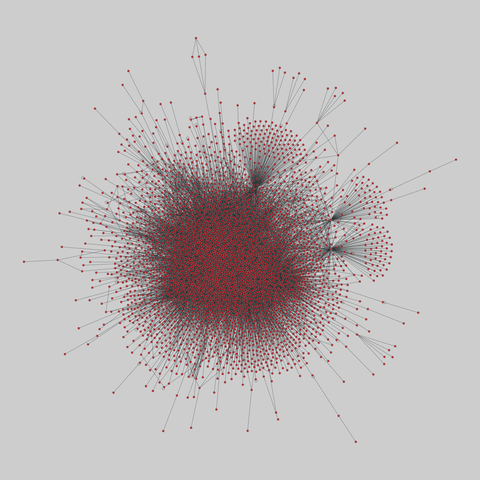
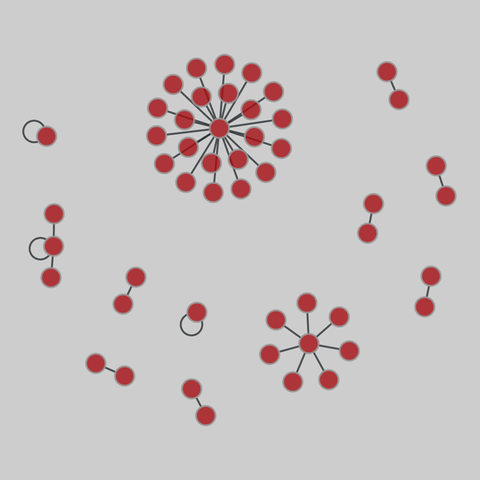
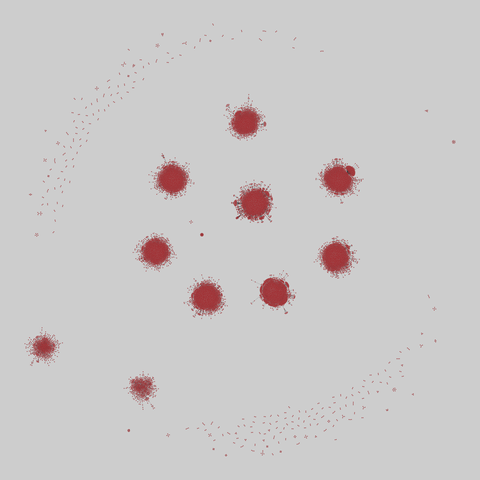
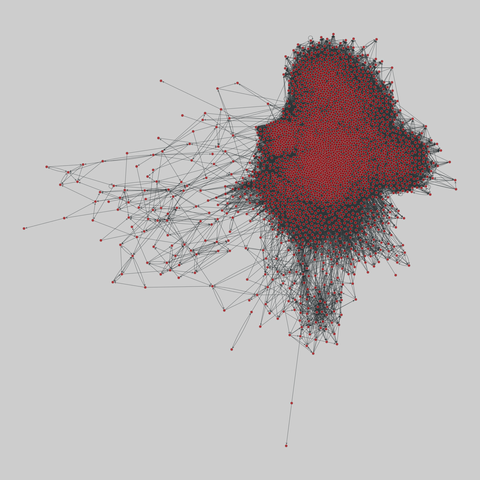
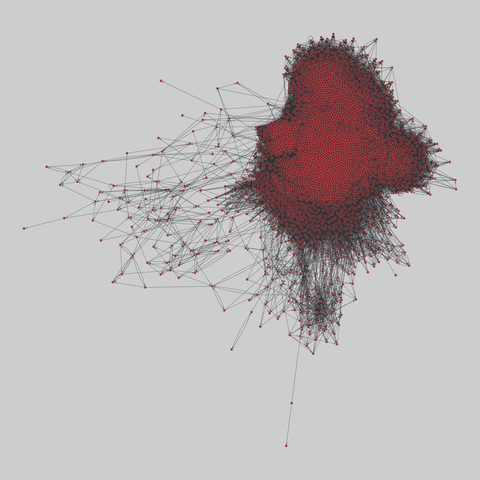
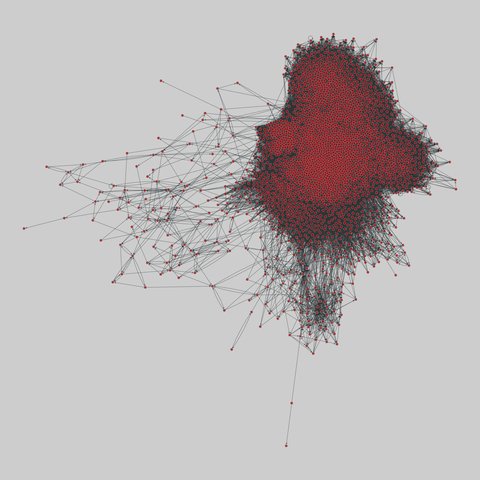
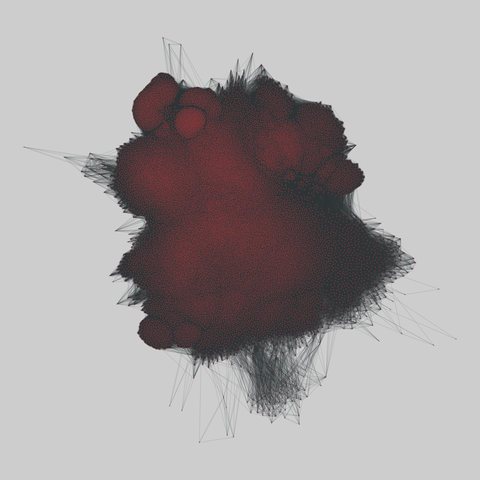
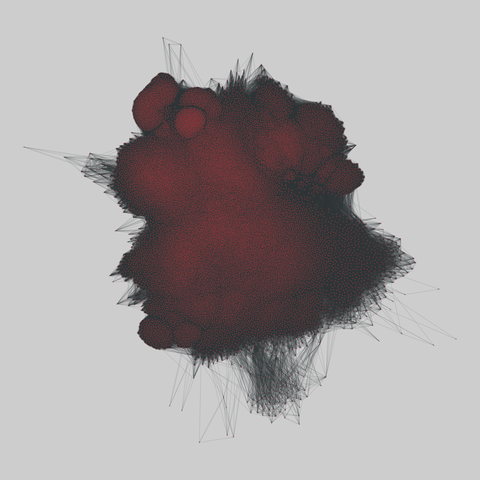
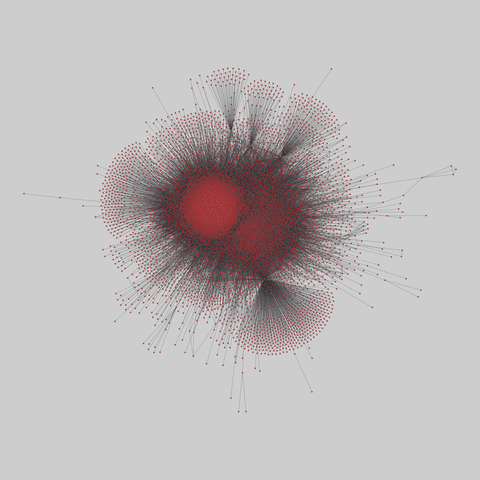
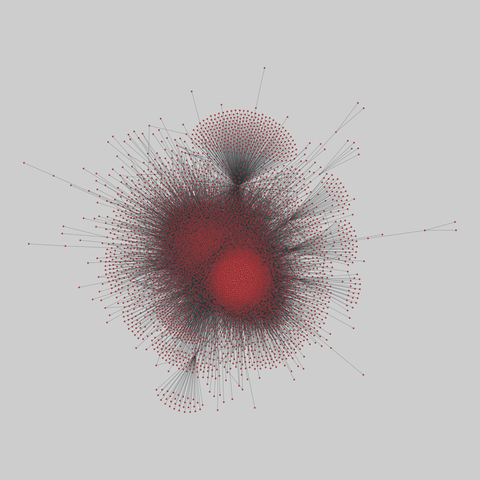
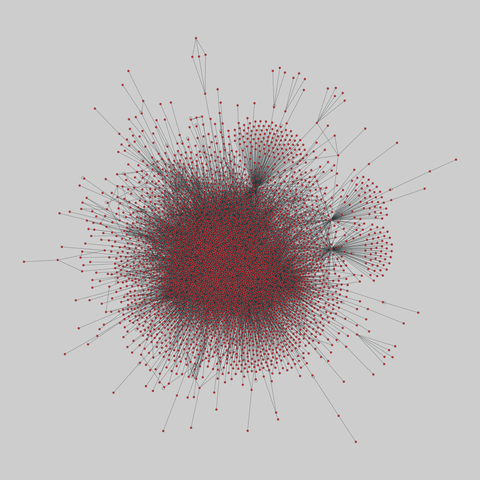
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
Parallel Neuron Groups in the Drosophila Brain
Robert Worden
https://arxiv.org/abs/2512.10525 https://arxiv.org/pdf/2512.10525 https://arxiv.org/html/2512.10525
arXiv:2512.10525v1 Announce Type: new
Abstract: The full connectome of an adult Drosophila enables a search for novel neural structures in the insect brain. I describe a new neural structure, called a Parallel Neuron Group (PNG). Two neurons are called parallel if they share a significant number of input neurons and output neurons. Most pairs of neurons in the Drosophila brain have very small parallel match. There are about twenty larger groups of neurons for which any pair of neurons in the group has a high match. These are the parallel groups. Parallel groups contain only about 1000 out of the 65,000 neurons in the brain, and have distinctive properties. There are groups in the right mushroom bodies, the antennal lobes, the lobula, and in two central neuropils (GNG and EB). Most parallel groups do not have lateral symmetry. A group usually has one major input neuron, which inputs to all the neurons in the group, and a small number of major output neurons. The major input and output neurons are laterally asymmetric. Parallel neuron groups present puzzles, such as: what does a group do, that could not be done by one larger neuron? Do all neurons in a group fire in synchrony, or do they perform different functions? Why are they laterally asymmetric? These may merit further investigation.
toXiv_bot_toot
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
fly_hemibrain: Fly hemibrain (2020)
A synaptic map of the hemibrain connectome of fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using EM reconstruction techniques. Neurons are labeled by their type. Edges are annotated by the connection strength between the neurons.
This network has 21739 nodes and 4259624 edges.
Tags: Biological, Connectome, Weighted, Metadata
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 3705 nodes and 15167 edges.
Tags: Biological, Protein interactions, Unweighted
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 50 nodes and 41 edges.
Tags: Biological, Protein interactions, Unweighted
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 27105 nodes and 334495 edges.
Tags: Biological, Protein interactions, Unweighted
Visual Function Profiles via Multi-Path Aggregation Reveal Neuron-Level Responses in the Drosophila Brain
Jiangping Xie, Ruohan Ren, Xiao Zhou, Ao Zheng, Jiasong Zhu, Wenyu Jiang, Ziran Zhao
https://arxiv.org/abs/2512.06934
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
fly_larva: Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`,…
fly_larva — Drosophila larva brain (2023)
A complete synaptic map of the brain connectome of the larva of the fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using three-dimensional electron microscopy–based reconstruction. Node metadata include the neuron hempisphere, hemispherical homologue, cell type, annotations, and inferred cluster. Edge metadata include the type of interaction (`'aa'`, `'ad'`, `'da'`, `'dd'`), and synapse count.
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 46404 nodes and 643319 edges.
Tags: Biological, Protein interactions, Unweighted
fly_hemibrain: Fly hemibrain (2020)
A synaptic map of the hemibrain connectome of fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using EM reconstruction techniques. Neurons are labeled by their type. Edges are annotated by the connection strength between the neurons.
This network has 21739 nodes and 4259624 edges.
Tags: Biological, Connectome, Weighted, Metadata
fly_hemibrain: Fly hemibrain (2020)
A synaptic map of the hemibrain connectome of fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using EM reconstruction techniques. Neurons are labeled by their type. Edges are annotated by the connection strength between the neurons.
This network has 21739 nodes and 4259624 edges.
Tags: Biological, Connectome, Weighted, Metadata
fly_hemibrain: Fly hemibrain (2020)
A synaptic map of the hemibrain connectome of fruit fly Drosophila melanogaster. Nodes are neurons, and edges are synaptic connections, traced individually from brain image sections using EM reconstruction techniques. Neurons are labeled by their type. Edges are annotated by the connection strength between the neurons.
This network has 21739 nodes and 4259624 edges.
Tags: Biological, Connectome, Weighted, Metadata
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 46404 nodes and 643319 edges.
Tags: Biological, Protein interactions, Unweighted
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 8699 nodes and 317814 edges.
Tags: Biological, Protein interactions, Unweighted
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 6618 nodes and 185239 edges.
Tags: Biological, Protein interactions, Unweighted
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 35355 nodes and 958936 edges.
Tags: Biological, Protein interactions, Unweighted
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 3705 nodes and 15167 edges.
Tags: Biological, Protein interactions, Unweighted