2026-01-10 06:00:05
mist: MIST protein interaction database (2020)
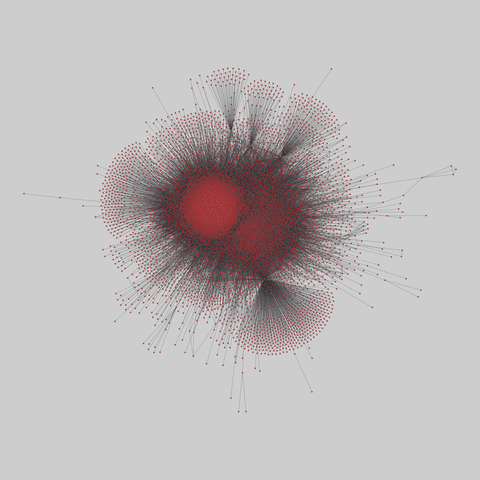
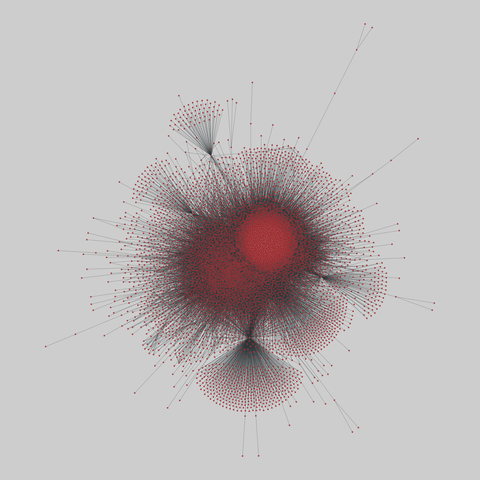
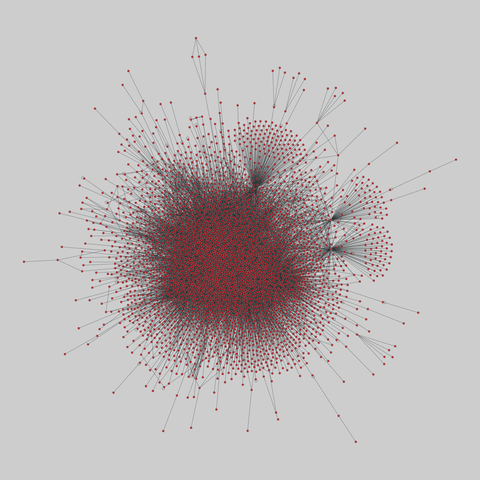
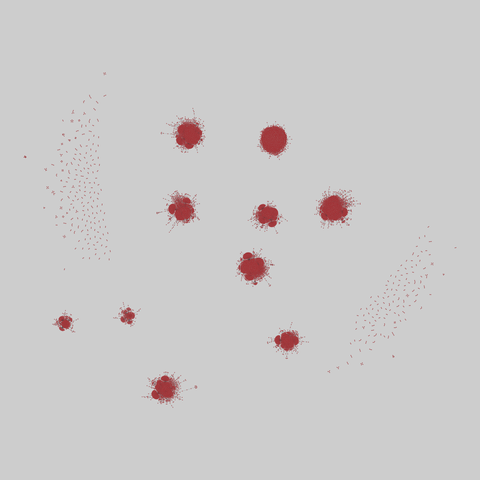
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 17845 nodes and 789779 edges.
Tags: Biological, Protein interactions, Unweighted