2025-07-18 09:25:22
Now out in #TMLR:
🍇 GRAPES: Learning to Sample Graphs for Scalable Graph Neural Networks 🍇
There's lots of work on sampling subgraphs for GNNs, but relatively little on making this sampling process _adaptive_. That is, learning to select the data from the graph that is relevant for your task.
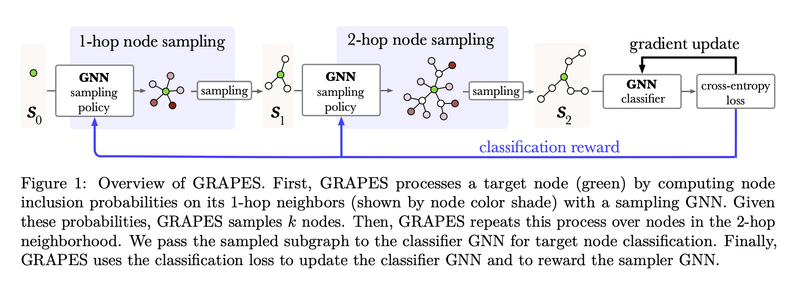
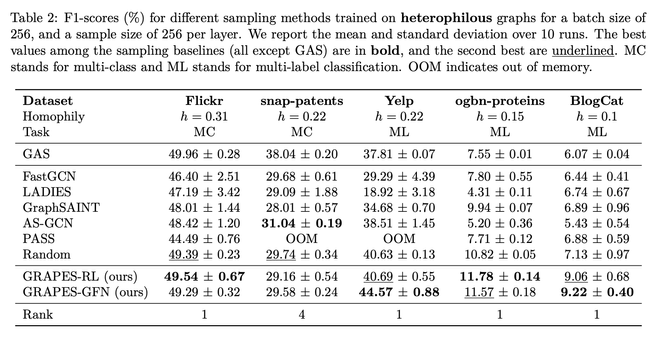
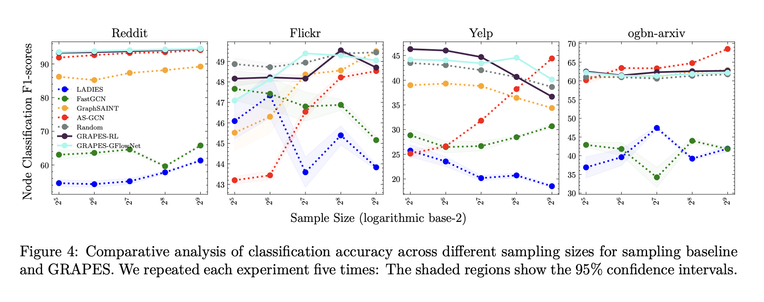
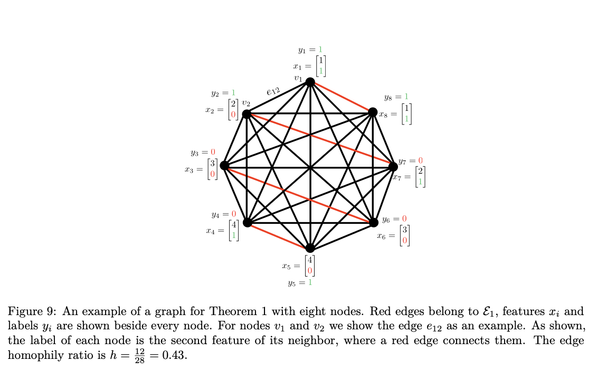
We introduce an RL-based and a GFLowNet-based sampler and show that the approach perf…