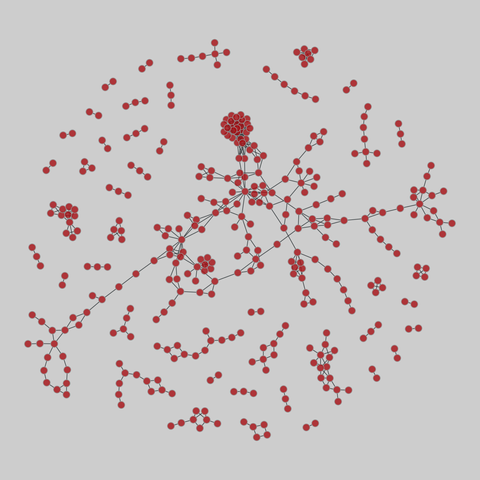
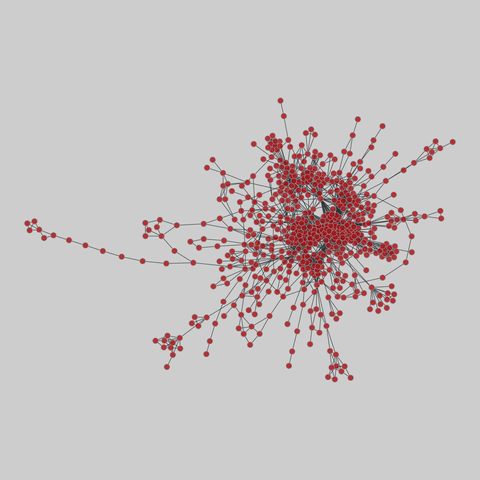
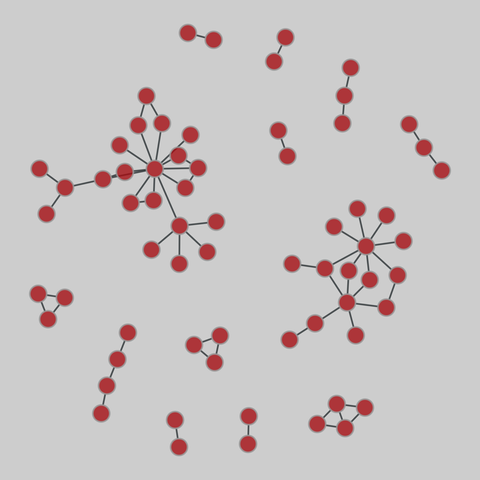
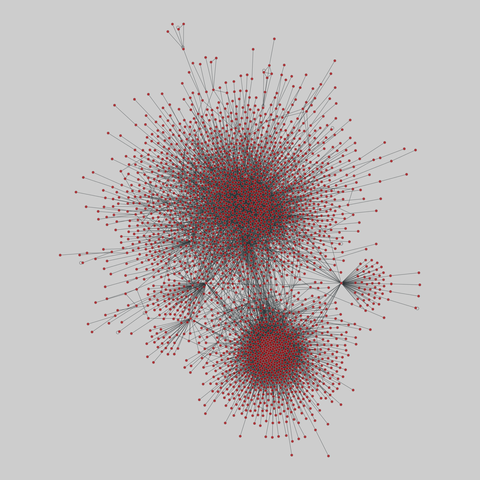
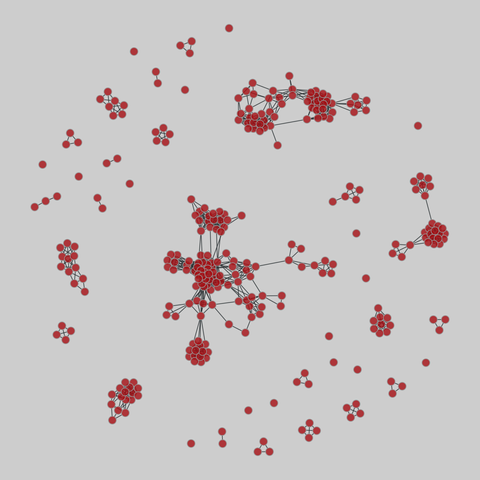
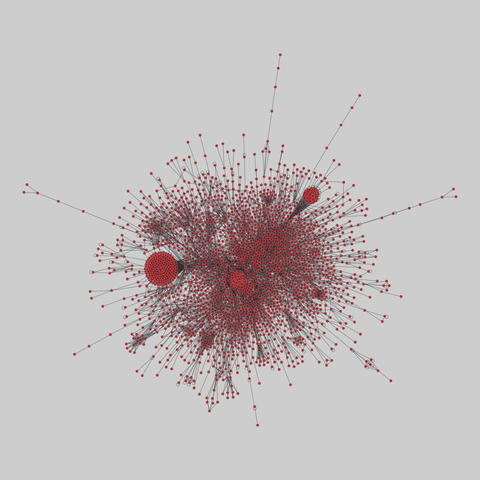
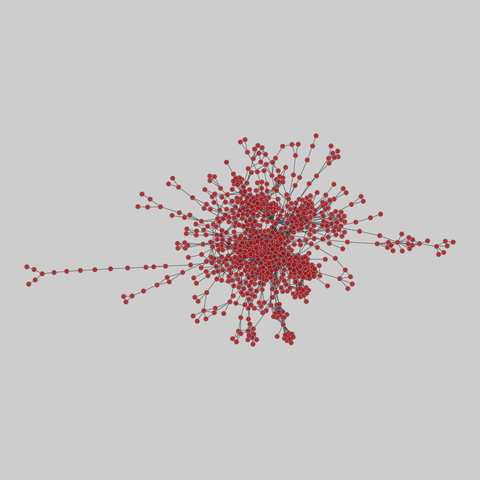
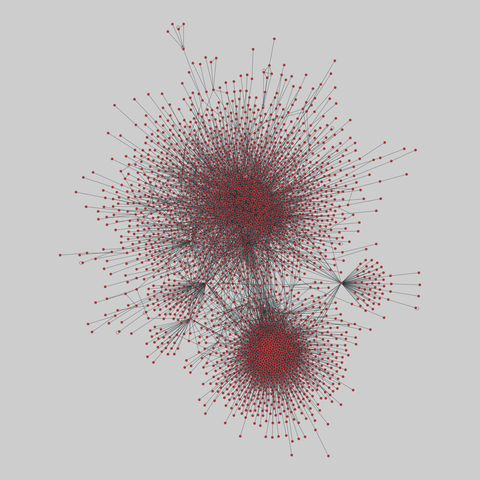
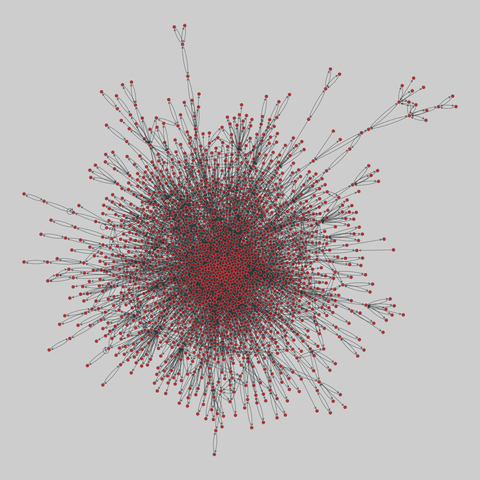
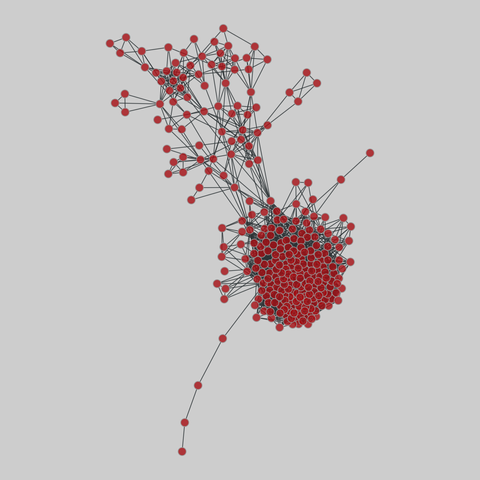
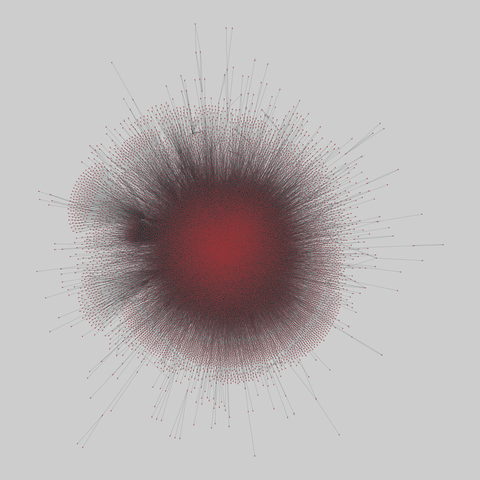
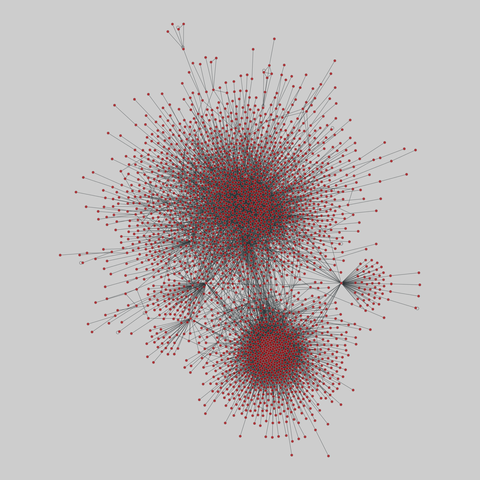
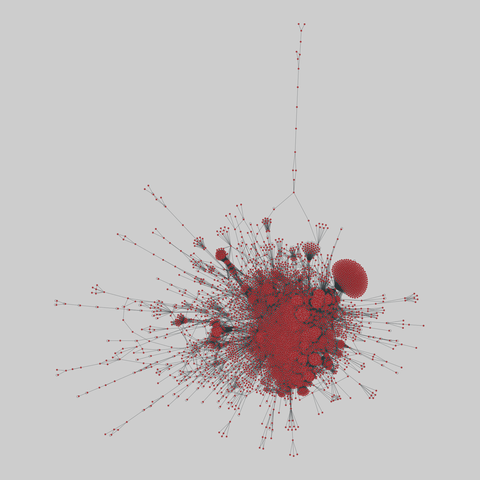
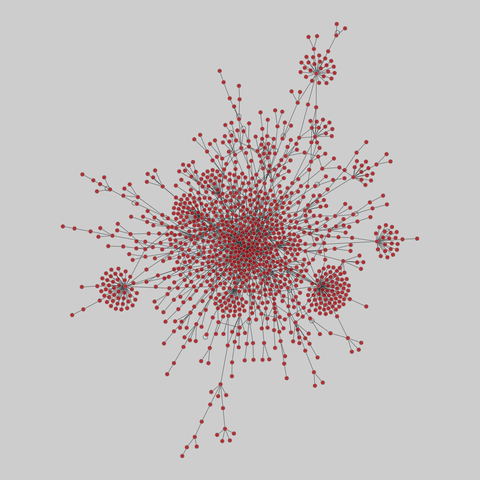
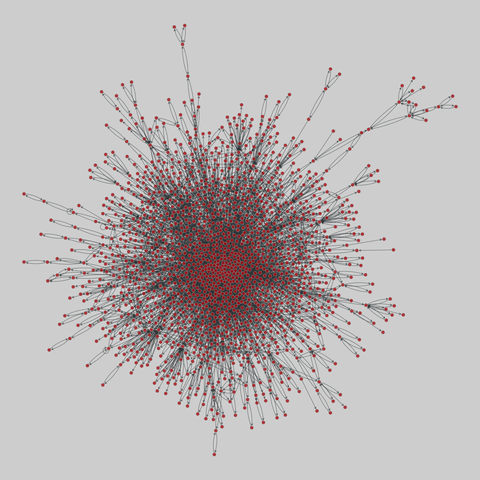
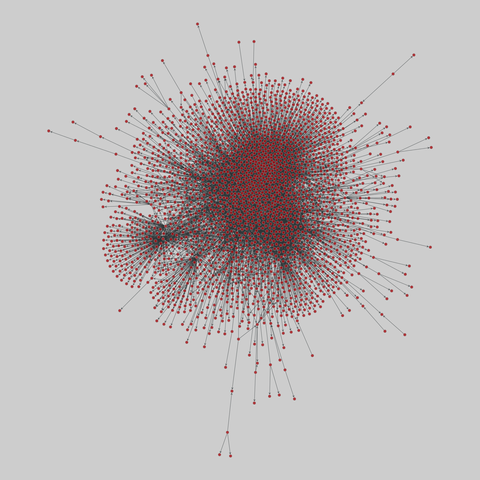
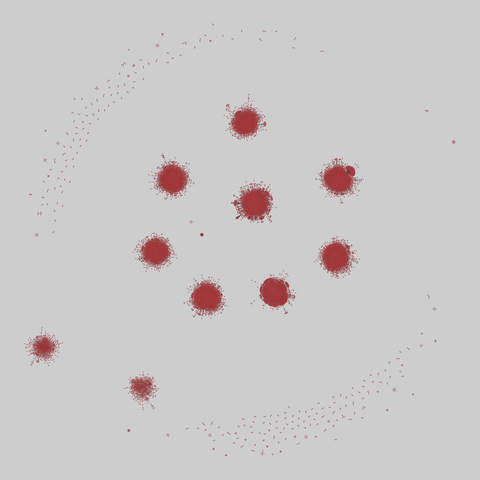
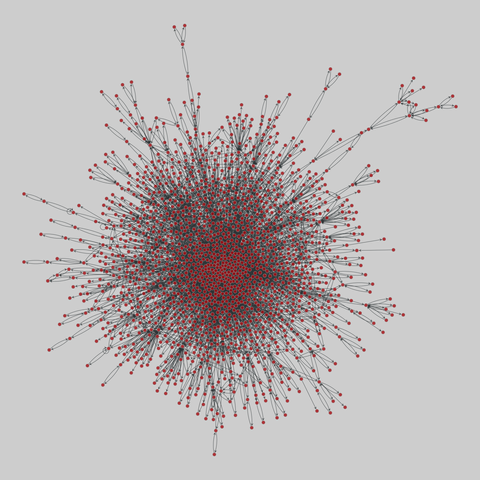
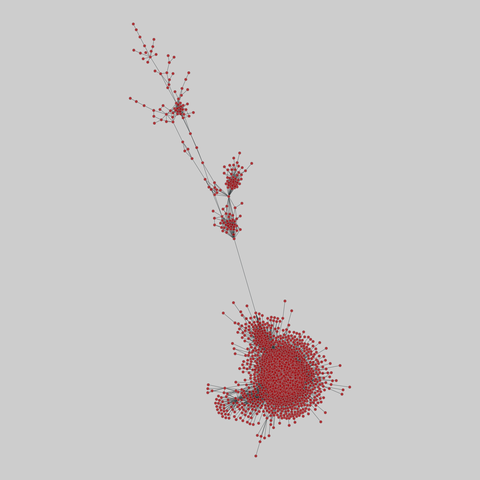
tree-of-life: Protein interactomes across the tree of life (2019)
Protein-protein iteraction networks ("interactome") for 1,840 species. An interactome captures all physical protein-protein interactions within one species, from direct biophysical protein-protein interactions to regulatory protein-DNA and metabolic interactions. .
This network has 374 nodes and 621 edges.
Tags: Biological, Protein interactions
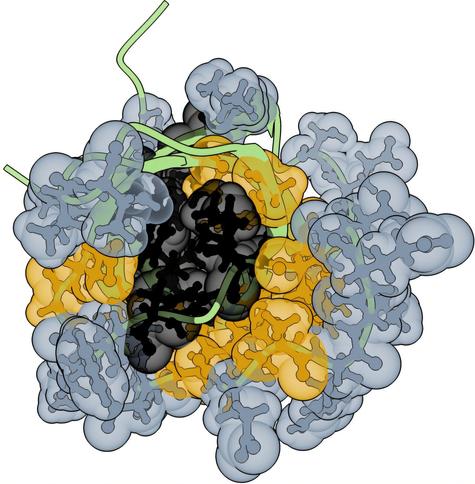
Learning Protein-Ligand Binding in Hyperbolic Space
Jianhui Wang, Wenyu Zhu, Bowen Gao, Xin Hong, Ya-Qin Zhang, Wei-Ying Ma, Yanyan Lan
https://arxiv.org/abs/2508.15480 https://…
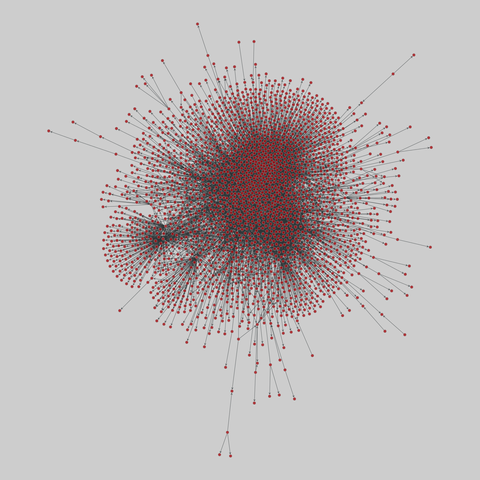
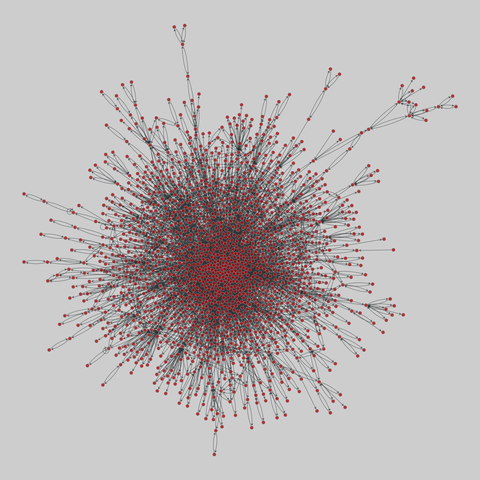
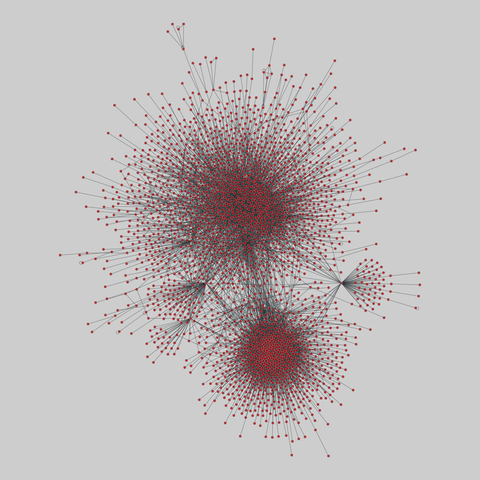
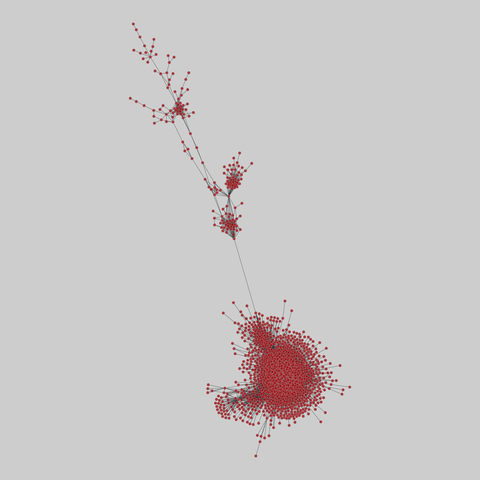
tree-of-life: Protein interactomes across the tree of life (2019)
Protein-protein iteraction networks ("interactome") for 1,840 species. An interactome captures all physical protein-protein interactions within one species, from direct biophysical protein-protein interactions to regulatory protein-DNA and metabolic interactions. .
This network has 780 nodes and 3099 edges.
Tags: Biological, Protein interactions
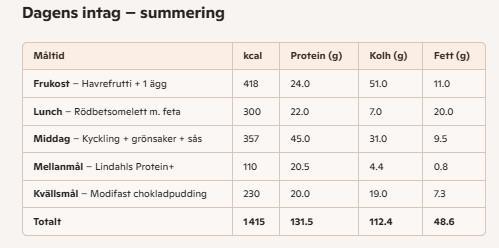
Så här kan en dag i mitt liv se ut just nu.
Chokladpuddingen på kvällen tillsammans med mitt ingefära-te är absolut nödvändigt, då varvar jag ner framför något på TV och slappar. Spetsar den med lakritspulver ibland för en extra egg.
Det är viktigt att inte ta ifrån sig själv allt livets goda. Något litet gott måste man få ha, och just den här puddingen är förvånansvärt god. Särskilt med lakrits :heart_fuchsia:
Barfing is their love language.♥️ A kinky male fruit fly barfs up a bit of his last meal and offers it as a nuptial meal to his intended. Discerning lady fruit flies swoon, and lap it up. #science #entomology
interactome_yeast: Coulomb yeast interactome (2005)
A network of protein-protein binding interactions among yeast proteins. Nodes represent proteins found in yeast (Saccharomyces cerevisiae) and an edge represents a binding interaction between two proteins.
This network has 1870 nodes and 2277 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_yeast: Coulomb yeast interactome (2005)
A network of protein-protein binding interactions among yeast proteins. Nodes represent proteins found in yeast (Saccharomyces cerevisiae) and an edge represents a binding interaction between two proteins.
This network has 1870 nodes and 2277 edges.
Tags: Biological, Protein interactions, Unweighted
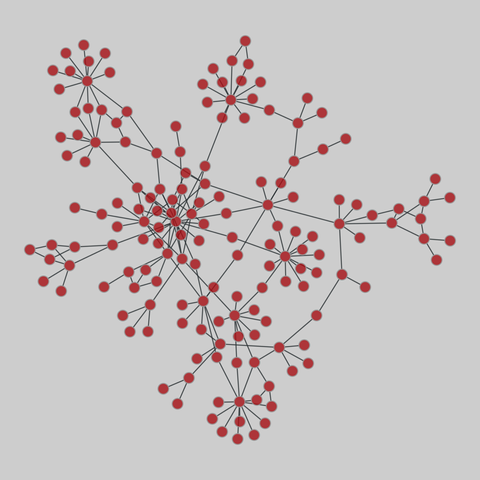
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 66 nodes and 65 edges.
Tags: Biological, Protein interactions, Unweighted
SHREC 2025: Protein surface shape retrieval including electrostatic potential
Taher Yacoub, Camille Depenveiller, Atsushi Tatsuma, Tin Barisin, Eugen Rusakov, Udo Gobel, Yuxu Peng, Shiqiang Deng, Yuki Kagaya, Joon Hong Park, Daisuke Kihara, Marco Guerra, Giorgio Palmieri, Andrea Ranieri, Ulderico Fugacci, Silvia Biasotti, Ruiwen He, Halim Benhabiles, Adnane Cabani, Karim Hammoudi, Haotian Li, Hao Huang, Chunyan Li, Alireza Tehrani, Fanwang Meng, Farnaz Heidar-Zadeh, Tuan-Anh Yang, Matt…
#Protein Powders and Shakes Contain High Levels of Lead. Also cadmium and inorganic arsenic.
Researchers at Consumer Reports tested various protein powders and shakes and their conclusion was Jesus Christ that's a lot of lead.
Crosslisted article(s) found for math.AT. https://arxiv.org/list/math.AT/new
[1/1]:
- Topological potentials guiding protein self-assembly
Ivan Spirandelli, Dmitriy Morozov, Arnur Nigmetov, Myfanwy E. Evans
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
Crosslisted article(s) found for cs.CG. https://arxiv.org/list/cs.CG/new
[1/1]:
- Topological potentials guiding protein self-assembly
Ivan Spirandelli, Dmitriy Morozov, Arnur Nigmetov, Myfanwy E. Evans
Decoding Translation-Related Functional Sequences in 5'UTRs Using Interpretable Deep Learning Models
Yuxi Lin, Yaxue Fang, Zehong Zhang, Zhouwu Liu, Siyun Zhong, Fulong Yu
https://arxiv.org/abs/2507.16801
Replaced article(s) found for cs.AI. https://arxiv.org/list/cs.AI/new
[2/3]:
- Uncertainty-Aware Adaptation of Large Language Models for Protein-Protein Interaction Analysis
Jantre, Wang, Park, Chopra, Jeon, Qian, Urban, Yoon
Replaced article(s) found for cs.CL. https://arxiv.org/list/cs.CL/new
[2/2]:
- Uncertainty-Aware Adaptation of Large Language Models for Protein-Protein Interaction Analysis
Jantre, Wang, Park, Chopra, Jeon, Qian, Urban, Yoon
Yeast growth is controlled by the proportional scaling of mRNA and ribosome concentrations
Xin Gao, Michael Lanz, Rosslyn Grosely, Jonas Cremer, Joseph Puglisi, Jan M. Skotheim
https://arxiv.org/abs/2508.14997
celegans_interactomes: C. elegans interactomes (2009)
Ten networks of protein-protein interactions in Caenorhabditis elegans (nematode), from yeast two-hybrid experiments, biological process maps, literature curation, orthologous interactions, and genetic interactions. The WI8 network combines WI2004, WI2007 and BPmaps, while the Integrated Network combines data from all sources.
This network has 431 nodes and 438 edges.
Tags: Biological, Protein interactions, Unweighted
Protein erobert die Kühlschränke vieler Konsumenten.
Adaptive Protein Design Protocols and Middleware
Aymen Alsaadi, Jonathan Ash, Mikhail Titov, Matteo Turilli, Andre Merzky, Shantenu Jha, Sagar Khare
https://arxiv.org/abs/2510.06396
Cooperation and competition of basepairing and electrostatic interactions in mixtures of DNA nanostars and polylysine
Gabrielle R. Abraham, Tianhao Li, Anna Nguyen, William M. Jacobs, Omar A. Saleh
https://arxiv.org/abs/2507.16179
Driving Accurate Allergen Prediction with Protein Language Models and Generalization-Focused Evaluation
Brian Shing-Hei Wong, Joshua Mincheol Kim, Sin-Hang Fung, Qing Xiong, Kelvin Fu-Kiu Ao, Junkang Wei, Ran Wang, Dan Michelle Wang, Jingying Zhou, Bo Feng, Alfred Sze-Lok Cheng, Kevin Y. Yip, Stephen Kwok-Wing Tsui, Qin Cao
https://arxiv.o…
Fast and Interpretable Protein Substructure Alignment via Optimal Transport
Zhiyu Wang, Bingxin Zhou, Jing Wang, Yang Tan, Weishu Zhao, Pietro Li\`o, Liang Hong
https://arxiv.org/abs/2510.11752
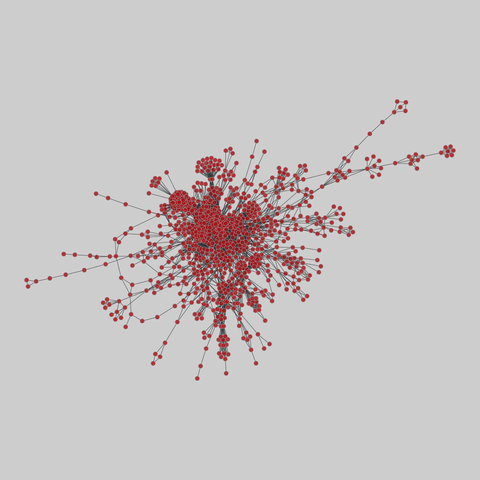
tree-of-life: Protein interactomes across the tree of life (2019)
Protein-protein iteraction networks ("interactome") for 1,840 species. An interactome captures all physical protein-protein interactions within one species, from direct biophysical protein-protein interactions to regulatory protein-DNA and metabolic interactions. .
This network has 1178 nodes and 7623 edges.
Tags: Biological, Protein interactions
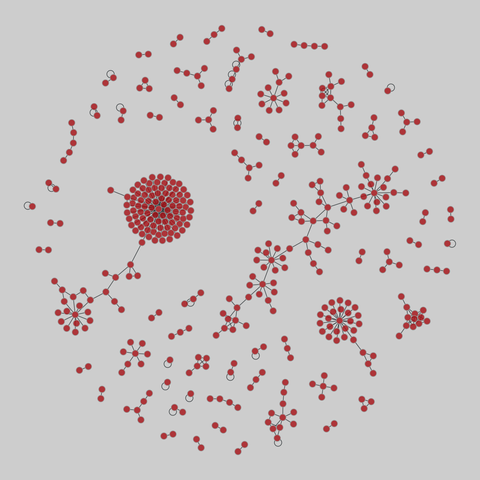
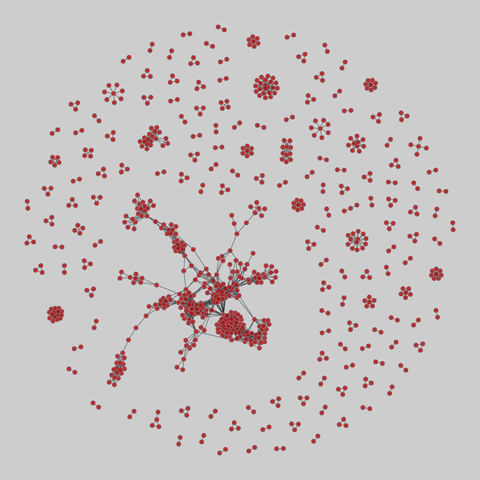
malaria_genes: Malaria var DBLa HVR networks
Networks of recombinant antigen genes from the human malaria parasite P. falciparum. Each of the 9 networks shares the same set of vertices but has different edges, corresponding to the 9 highly variable regions (HVRs) in the DBLa domain of the var protein. Nodes are var genes, and two genes are connected if they share a substring whose length is statistically significant. Metadata includes two types of node labels, both based on sequence st…
Replaced article(s) found for q-bio.BM. https://arxiv.org/list/q-bio.BM/new
[1/1]:
- Protein folding classes -- High-dimensional geometry of amino acid composition space revisited
Boryeu Mao
Virtuous Machines: Towards Artificial General Science
Gabrielle Wehr, Reuben Rideaux, Amaya J. Fox, David R. Lightfoot, Jason Tangen, Jason B. Mattingley, Shane E. Ehrhardt
https://arxiv.org/abs/2508.13421
Energy-Based Models for Predicting Mutational Effects on Proteins
Patrick Soga, Zhenyu Lei, Yinhan He, Camille Bilodeau, Jundong Li
https://arxiv.org/abs/2508.10629 https://
celegans_interactomes: C. elegans interactomes (2009)
Ten networks of protein-protein interactions in Caenorhabditis elegans (nematode), from yeast two-hybrid experiments, biological process maps, literature curation, orthologous interactions, and genetic interactions. The WI8 network combines WI2004, WI2007 and BPmaps, while the Integrated Network combines data from all sources.
This network has 2724 nodes and 14322 edges.
Tags: Biological, Protein interactions, Unweighted…
tree-of-life: Protein interactomes across the tree of life (2019)
Protein-protein iteraction networks ("interactome") for 1,840 species. An interactome captures all physical protein-protein interactions within one species, from direct biophysical protein-protein interactions to regulatory protein-DNA and metabolic interactions. .
This network has 543 nodes and 1491 edges.
Tags: Biological, Protein interactions
Crosslisted article(s) found for q-bio.BM. https://arxiv.org/list/q-bio.BM/new
[1/1]:
- Monte Carlo Tree Diffusion with Multiple Experts for Protein Design
Liu, Cao, Jiang, Luo, Duan, Wang, Sosnick, Xu, Stevens
tree-of-life: Protein interactomes across the tree of life (2019)
Protein-protein iteraction networks ("interactome") for 1,840 species. An interactome captures all physical protein-protein interactions within one species, from direct biophysical protein-protein interactions to regulatory protein-DNA and metabolic interactions. .
This network has 971 nodes and 4170 edges.
Tags: Biological, Protein interactions
Multimodal Quantum Vision Transformer for Enzyme Commission Classification from Biochemical Representations
Murat Isik, Mandeep Kaur Saggi, Humaira Gowher, Sabre Kais
https://arxiv.org/abs/2508.14844
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
Synthetic Protein-Ligand Complex Generation for Deep Molecular Docking
Sofiene Khiari, Matthew R. Masters, Amr H. Mahmoud, Markus A. Lill
https://arxiv.org/abs/2509.12915 https:…
PRISM: Enhancing Protein Inverse Folding through Fine-Grained Retrieval on Structure-Sequence Multimodal Representations
Sazan Mahbub, Souvik Kundu, Eric P. Xing
https://arxiv.org/abs/2510.11750
A warning: y’all know I like #startups and I like #foodscience and I’m an adventurous eater and no complainer, but Wilde Protein Chips are one of the worst products I’ve ever tried — I actually bit into the included silica gel packet and it wasn’t too different from the chips.
interactome_pdz: PDZ-domain interactome (2005)
A network of PDZ-domain-mediated protein–protein binding interactions, extracted from the PDZBase database. Nodes represent proteins and an edge represents a binding interaction between two proteins.
This network has 212 nodes and 244 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_figeys: Figeys human interactome (2007)
A network of human proteins and their binding interactions. Nodes represent proteins and an edge represents an interaction between two proteins, as inferred using a mass spectrometry‐based approach.
This network has 2239 nodes and 6452 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_stelzl: Stelzl human interactome (2005)
A network of human proteins and their binding interactions. Nodes represent proteins and an edge represents an interaction between two proteins, as inferred via high-throughput Y2H experiments using bait and prey methodology.
This network has 1706 nodes and 6207 edges.
Tags: Biological, Protein interactions, Unweighted
Evolutionary Profiles for Protein Fitness Prediction
Jigang Fan, Xiaoran Jiao, Shengdong Lin, Zhanming Liang, Weian Mao, Chenchen Jing, Hao Chen, Chunhua Shen
https://arxiv.org/abs/2510.07286
interactome_stelzl: Stelzl human interactome (2005)
A network of human proteins and their binding interactions. Nodes represent proteins and an edge represents an interaction between two proteins, as inferred via high-throughput Y2H experiments using bait and prey methodology.
This network has 1706 nodes and 6207 edges.
Tags: Biological, Protein interactions, Unweighted
malaria_genes: Malaria var DBLa HVR networks
Networks of recombinant antigen genes from the human malaria parasite P. falciparum. Each of the 9 networks shares the same set of vertices but has different edges, corresponding to the 9 highly variable regions (HVRs) in the DBLa domain of the var protein. Nodes are var genes, and two genes are connected if they share a substring whose length is statistically significant. Metadata includes two types of node labels, both based on sequence st…
genetic_multiplex: Multiplex genetic interactions (2014)
Multiplex networks representing different types of genetic interactions, for different organisms. Layers represent (i) physical, (ii) association, (iii) co-localization, (iv) direct, and (v) suppressive, (vi) additive or synthetic genetic interaction. Edge direction (i,j) indicates gene i interacting with gene j.
This network has 18222 nodes and 170899 edges.
Tags: Biological, Gene regulation, Protein interactions, Un…
Deep Learning-based QSAR Model for Therapeutic Strategies Targeting SmTGR Protein's Immune Modulating Role in Host-Parasite Interaction
Belaguppa Manjunath Ashwin Desai, Belaguppa Manjunath Anirudh, Kalyani S Biju, Vondhana Ramesh, Pronama Biswas
https://arxiv.org/abs/2508.12653
ProteoKnight: Convolution-based phage virion protein classification and uncertainty analysis
Samiha Afaf Neha, Abir Ahammed Bhuiyan, Md. Ishrak Khan
https://arxiv.org/abs/2508.07345
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
collins_yeast: Collins yeast interactome (2007)
Network of protein-protein interactions in Saccharomyces cerevisiae (budding yeast), measured by co-complex associations identified by high-throughput affinity purification and mass spectrometry (AP/MS).
This network has 1622 nodes and 9070 edges.
Tags: Biological, Protein interactions, Unweighted
IBEX: Information-Bottleneck-EXplored Coarse-to-Fine Molecular Generation under Limited Data
Dong Xu, Zhangfan Yang, Jenna Xinyi Yao, Shuangbao Song, Zexuan Zhu, Junkai Ji
https://arxiv.org/abs/2508.10775
tree-of-life: Protein interactomes across the tree of life (2019)
Protein-protein iteraction networks ("interactome") for 1,840 species. An interactome captures all physical protein-protein interactions within one species, from direct biophysical protein-protein interactions to regulatory protein-DNA and metabolic interactions. .
This network has 813 nodes and 2611 edges.
Tags: Biological, Protein interactions
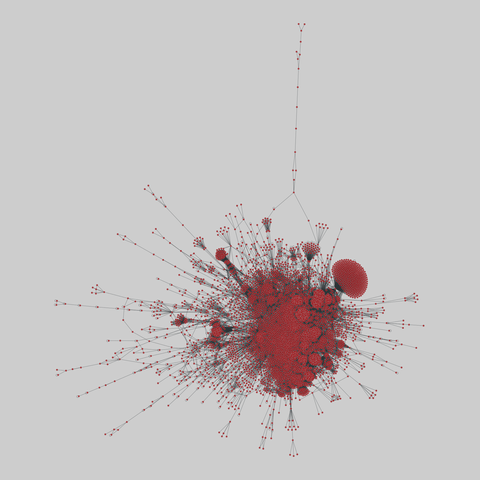
reactome: Joshi-Tope human protein interactome (2005)
A network of human proteins and their binding interactions, extracted from Reactome project. Nodes represent proteins and an edge represents a binding interaction between two proteins.
This network has 6327 nodes and 147547 edges.
Tags: Biological, Protein interactions, Unweighted
https:/…
tree-of-life: Protein interactomes across the tree of life (2019)
Protein-protein iteraction networks ("interactome") for 1,840 species. An interactome captures all physical protein-protein interactions within one species, from direct biophysical protein-protein interactions to regulatory protein-DNA and metabolic interactions. .
This network has 1501 nodes and 6848 edges.
Tags: Biological, Protein interactions
interactome_yeast: Coulomb yeast interactome (2005)
A network of protein-protein binding interactions among yeast proteins. Nodes represent proteins found in yeast (Saccharomyces cerevisiae) and an edge represents a binding interaction between two proteins.
This network has 1870 nodes and 2277 edges.
Tags: Biological, Protein interactions, Unweighted
malaria_genes: Malaria var DBLa HVR networks
Networks of recombinant antigen genes from the human malaria parasite P. falciparum. Each of the 9 networks shares the same set of vertices but has different edges, corresponding to the 9 highly variable regions (HVRs) in the DBLa domain of the var protein. Nodes are var genes, and two genes are connected if they share a substring whose length is statistically significant. Metadata includes two types of node labels, both based on sequence st…
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
tree-of-life: Protein interactomes across the tree of life (2019)
Protein-protein iteraction networks ("interactome") for 1,840 species. An interactome captures all physical protein-protein interactions within one species, from direct biophysical protein-protein interactions to regulatory protein-DNA and metabolic interactions. .
This network has 277 nodes and 291 edges.
Tags: Biological, Protein interactions
collins_yeast: Collins yeast interactome (2007)
Network of protein-protein interactions in Saccharomyces cerevisiae (budding yeast), measured by co-complex associations identified by high-throughput affinity purification and mass spectrometry (AP/MS).
This network has 1622 nodes and 9070 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_yeast: Coulomb yeast interactome (2005)
A network of protein-protein binding interactions among yeast proteins. Nodes represent proteins found in yeast (Saccharomyces cerevisiae) and an edge represents a binding interaction between two proteins.
This network has 1870 nodes and 2277 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_yeast: Coulomb yeast interactome (2005)
A network of protein-protein binding interactions among yeast proteins. Nodes represent proteins found in yeast (Saccharomyces cerevisiae) and an edge represents a binding interaction between two proteins.
This network has 1870 nodes and 2277 edges.
Tags: Biological, Protein interactions, Unweighted
reactome: Joshi-Tope human protein interactome (2005)
A network of human proteins and their binding interactions, extracted from Reactome project. Nodes represent proteins and an edge represents a binding interaction between two proteins.
This network has 6327 nodes and 147547 edges.
Tags: Biological, Protein interactions, Unweighted
https:/…
collins_yeast: Collins yeast interactome (2007)
Network of protein-protein interactions in Saccharomyces cerevisiae (budding yeast), measured by co-complex associations identified by high-throughput affinity purification and mass spectrometry (AP/MS).
This network has 1622 nodes and 9070 edges.
Tags: Biological, Protein interactions, Unweighted
celegans_interactomes: C. elegans interactomes (2009)
Ten networks of protein-protein interactions in Caenorhabditis elegans (nematode), from yeast two-hybrid experiments, biological process maps, literature curation, orthologous interactions, and genetic interactions. The WI8 network combines WI2004, WI2007 and BPmaps, while the Integrated Network combines data from all sources.
This network has 2724 nodes and 14322 edges.
Tags: Biological, Protein interactions, Unweighted…
celegans_interactomes: C. elegans interactomes (2009)
Ten networks of protein-protein interactions in Caenorhabditis elegans (nematode), from yeast two-hybrid experiments, biological process maps, literature curation, orthologous interactions, and genetic interactions. The WI8 network combines WI2004, WI2007 and BPmaps, while the Integrated Network combines data from all sources.
This network has 2436 nodes and 136930 edges.
Tags: Biological, Protein interactions, Unweighte…
celegans_interactomes: C. elegans interactomes (2009)
Ten networks of protein-protein interactions in Caenorhabditis elegans (nematode), from yeast two-hybrid experiments, biological process maps, literature curation, orthologous interactions, and genetic interactions. The WI8 network combines WI2004, WI2007 and BPmaps, while the Integrated Network combines data from all sources.
This network has 1496 nodes and 1816 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_stelzl: Stelzl human interactome (2005)
A network of human proteins and their binding interactions. Nodes represent proteins and an edge represents an interaction between two proteins, as inferred via high-throughput Y2H experiments using bait and prey methodology.
This network has 1706 nodes and 6207 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_stelzl: Stelzl human interactome (2005)
A network of human proteins and their binding interactions. Nodes represent proteins and an edge represents an interaction between two proteins, as inferred via high-throughput Y2H experiments using bait and prey methodology.
This network has 1706 nodes and 6207 edges.
Tags: Biological, Protein interactions, Unweighted
drosophila_flybi: Fruit fly protein interactions (Drosophila melanogaster)
Binary protein-protein interactions (PPIs) for Drosophila melanogaster, containing 8723 PPIs among 2939 proteins. The iteractions were identified using a yeast two-hybrid (Y2H) analysis.
This network has 2939 nodes and 8723 edges.
Tags: Biological, Protein interactions, Unweighted
celegans_interactomes: C. elegans interactomes (2009)
Ten networks of protein-protein interactions in Caenorhabditis elegans (nematode), from yeast two-hybrid experiments, biological process maps, literature curation, orthologous interactions, and genetic interactions. The WI8 network combines WI2004, WI2007 and BPmaps, while the Integrated Network combines data from all sources.
This network has 912 nodes and 22738 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_yeast: Coulomb yeast interactome (2005)
A network of protein-protein binding interactions among yeast proteins. Nodes represent proteins found in yeast (Saccharomyces cerevisiae) and an edge represents a binding interaction between two proteins.
This network has 1870 nodes and 2277 edges.
Tags: Biological, Protein interactions, Unweighted
reactome: Joshi-Tope human protein interactome (2005)
A network of human proteins and their binding interactions, extracted from Reactome project. Nodes represent proteins and an edge represents a binding interaction between two proteins.
This network has 6327 nodes and 147547 edges.
Tags: Biological, Protein interactions, Unweighted
https:/…
celegans_interactomes: C. elegans interactomes (2009)
Ten networks of protein-protein interactions in Caenorhabditis elegans (nematode), from yeast two-hybrid experiments, biological process maps, literature curation, orthologous interactions, and genetic interactions. The WI8 network combines WI2004, WI2007 and BPmaps, while the Integrated Network combines data from all sources.
This network has 759 nodes and 1593 edges.
Tags: Biological, Protein interactions, Unweighted…
interactome_figeys: Figeys human interactome (2007)
A network of human proteins and their binding interactions. Nodes represent proteins and an edge represents an interaction between two proteins, as inferred using a mass spectrometry‐based approach.
This network has 2239 nodes and 6452 edges.
Tags: Biological, Protein interactions, Unweighted
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 27105 nodes and 334495 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_stelzl: Stelzl human interactome (2005)
A network of human proteins and their binding interactions. Nodes represent proteins and an edge represents an interaction between two proteins, as inferred via high-throughput Y2H experiments using bait and prey methodology.
This network has 1706 nodes and 6207 edges.
Tags: Biological, Protein interactions, Unweighted
mist: MIST protein interaction database (2020)
The Molecular Interaction Search Tool (MIST) is a comprehensive resource of molecular interactions, assembled from severla primary sources. MIST currently supports several species, including:.
This network has 46404 nodes and 643319 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_yeast: Coulomb yeast interactome (2005)
A network of protein-protein binding interactions among yeast proteins. Nodes represent proteins found in yeast (Saccharomyces cerevisiae) and an edge represents a binding interaction between two proteins.
This network has 1870 nodes and 2277 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_yeast: Coulomb yeast interactome (2005)
A network of protein-protein binding interactions among yeast proteins. Nodes represent proteins found in yeast (Saccharomyces cerevisiae) and an edge represents a binding interaction between two proteins.
This network has 1870 nodes and 2277 edges.
Tags: Biological, Protein interactions, Unweighted
collins_yeast: Collins yeast interactome (2007)
Network of protein-protein interactions in Saccharomyces cerevisiae (budding yeast), measured by co-complex associations identified by high-throughput affinity purification and mass spectrometry (AP/MS).
This network has 1622 nodes and 9070 edges.
Tags: Biological, Protein interactions, Unweighted
interactome_yeast: Coulomb yeast interactome (2005)
A network of protein-protein binding interactions among yeast proteins. Nodes represent proteins found in yeast (Saccharomyces cerevisiae) and an edge represents a binding interaction between two proteins.
This network has 1870 nodes and 2277 edges.
Tags: Biological, Protein interactions, Unweighted
celegans_interactomes: C. elegans interactomes (2009)
Ten networks of protein-protein interactions in Caenorhabditis elegans (nematode), from yeast two-hybrid experiments, biological process maps, literature curation, orthologous interactions, and genetic interactions. The WI8 network combines WI2004, WI2007 and BPmaps, while the Integrated Network combines data from all sources.
This network has 2436 nodes and 136930 edges.
Tags: Biological, Protein interactions, Unweighte…